Description of result pages
For this motif data base, we've slightly changed the typical HOMERv3 layout. Additional description of the typical contentof results pages is provided below. Available information will appear once you point your mouse over column headers.
Known motif results will look like this:
Total target sequences = 33960
Total background sequences = 33914Rank | Motif |
Name | P-value | q-value(Benjamini) | Enrichment | Motif File |
PDF |
1
| AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq/Homer | 1e-1217 | 0.0000 | 8524.0 (25.10%) targets3625.4 (10.69%) backgr. | motif file (matrix) | ||
2
| HIF1b(HLH)/O785-HIF1b-ChIP-Seq(GSE34871)/Homer | 1e-1215 | 0.0000 | 7492.0 (22.06%) targets2944.0 (8.68%) backgr. | motif file (matrix) | ||
3
| Jun-AP1(bZIP)/K562-cJun-ChIP-Seq/Homer | 1e-1022 | 0.0000 | 3843.0 (11.32%) targets1036.1 (3.06%) backgr. | motif file (matrix) |
De novo motif results will look like this:
Total target sequences = 33960
Total background sequences = 33914Rank | Motif | P-value | Enrichment | Best Match and Details |
1
| 1e-1333 | 7265.0 (21.39%) targets 2647.8 (7.81%) backgr. | HIF1b(HLH)/O785-HIF1b-ChIP-Seq(GSE34871)/Homer (0.99) More Info/Top 5 Known Motifs | |
2
| 1e-840 | 11031.0 (32.48%) targets 6230.1 (18.38%) backgr. | ATF3_f1_HM09 (0.88) More Info/Top 5 Known Motifs | |
3
| 1e-694 | 10607.0 (31.24%) targets 6263.0 (18.48%) backgr. | MA0080.2_SPI1 (0.95) More Info/Top 5 Known Motifs |
Additional information and matches to known motifs (will open by clicking the 'More Info/Top 5 motifs' link)
Detailed information for motif1
| P-value: | 1e-1333 |
| ln P-value: | -3.070e+03 |
| Information content per bp: | 1.608 |
| Number of target sequences with motif: | 7265.0 |
| Percentage of target sequences with motif: | 21.39% |
| Number of background sequences with motif: | 2647.8 |
| Percentage of background sequences with motif: | 7.81% |
| Average position of motif in targets: | 201.8 +/- 76.8bp |
| Average position of motif in background: | 196.8 +/- 116.6bp |
| Strand bias (log2 ratio + to - strand density): | 0.0 |
| Multiplicity (# of sites on avg that occur together): | 1.17 |
| Motif file: | file (matrix) reverse opposite |
| PDF format logos: | file (matrix) reverse opposite |
Motif logos
| Forward: |
| Reverse: |
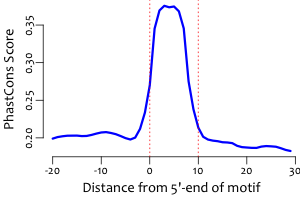
Conservation across vertebrates
Matches to known motifs
HIF1b(HLH)/O785-HIF1b-ChIP-Seq(GSE34871)/Homer
| ||||||||||||||
Jun-AP1(bZIP)/K562-cJun-ChIP-Seq/Homer
| ||||||||||||||
AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq/Homer
| ||||||||||||||
MA0303.1_GCN4
| ||||||||||||||
MA0099.1_Fos
|

